Introduction
Large language models (LLMs) are increasingly considered for use in healthcare. Although no commercially available LLMs are currently approved by the FDA for use in medical decision support settings1, top-performing LLMs like GPT-4o, Claude, and Med-PaLM have nonetheless demonstrated superior performance over clinicians on medical exams like the US Medical Licensing Exam (USMLE)2,3,4. LLMs have already made their way into patient care today, from being used as chatbots for mental health therapy5,6 to users finding diagnoses for uncommon diseases that physicians missed7. A growing number of clinicians report using LLMs in their clinical practice or education8,9.
However, LLMs are prone to hallucination, where the model generates statements not backed by any source10,11,12. Particularly in the medical domain, this can erode user trust and potentially harm patients by providing erroneous advice13,14 or discriminating based on patient backgrounds15. Lack of trust is commonly cited as the number one deterrent against clinicians adopting LLMs in their clinical practice16,17, and in particular, the inability of LLMs to generate supporting sources for medical statements in their responses18.
The need to cite the sources for medical statements goes beyond gaining clinician and patient trust -- there is also an urgent regulatory case as well19. The US Food and Drug Administration (FDA) has repeatedly called for regulating LLMs used as decision support tools20,21. Assessing the degree to which LLMs reliably convey existing, trustworthy medical knowledge is important for informing future regulatory frameworks regarding medical LLMs.
LLMs should be capable of reliably providing relevant sources to allow users and regulators to audit the reliability of their statements. Recent advancements in LLM capabilities (e.g., improved instruction fine-tuning) have enabled models to routinely provide sources upon request. Retrieval augmented generation (RAG), in particular, allows models to perform real-time searches for web references relevant to the query. However, even if the references are from valid and legitimate websites, it is still unclear the extent to which these provided sources contain content that actually supports the claims made in the model’s generated responses.
To address these important challenges, this paper makes the following contributions. First, given the costly nature of high-quality medical expert annotations and the rapid pace of ongoing development in LLMs, we propose an automated evaluation framework, called SourceCheckup, to create medical questions and to score how well LLMs can provide relevant sources in their answers to these questions. We verify that this framework is highly accurate, finding 89% agreement with a consensus of three US-licensed medical experts and a higher rate of agreement than any pair of medical experts. Second, we evaluate top-performing, commercially available LLMs (GPT-4o (RAG and API), Claude v2.1, Mistral Medium, and Gemini (RAG and API)) and find that models without access to the Web only produce valid URLs between 40% to 70% of the time. Retrieval-augmented generation (RAG)-enabled GPT-4o and Gemini Ultra 1.0, which have search engine access, do not suffer from URL hallucination, but still fail to produce references that support all the statements in the response nearly half of the time. Finally, we open source our dataset of 800 medical questions created from webpages pulled from the Mayo Clinic and Reddit, as well as a clinician-annotated subset of 400 question/answer pairs. Our findings highlight an important gap in the viability of LLMs for clinical medicine and have crucial implications for the medical adoption of LLMs.
Related works
There is a growing body of work on measuring and improving source attribution in language models22,23,24. Benchmark datasets introduced in works such as WebGPT25, ExpertQA26, WebCPM27, and HAGRID28 aggregate open-domain subjects from web pages like Wikipedia in a question-answer format. However, the evaluations of these datasets were performed by manual human verification, which can be costly and time-intensive29 and difficult to replicate.
There have been several works recently that have demonstrated the usefulness of using language models themselves in automatically scoring source attribution from LLMs. For instance, ALCE24, AttributedQA30, and GopherCite31 use supervised language models to perform automated evaluation of LLMs. More relevantly, given the advent of powerful instruction-fine-tuned LLMs, FactScore32 and AttrScore33 demonstrate that ChatGPT can be used as a useful evaluator of source attribution, but ChatGPT itself performs poorly when evaluated for source attribution34,35.
Our proposed method makes three contributions. First, we construct a dedicated corpus of medical-specific statement-source pairs (nearly 58 K examples from over 800 reference documents). Second, we provide evidence that GPT-4o is a highly effective evaluator of source attribution in the medical domain by showing strong agreement with a panel of three US-licensed medical doctors. Third, we use our automated framework to evaluate seven state-of-the-art, commercially available LLMs commonly used by patients and clinicians today.
Results
Question generation and response parsing
Both medical doctors tasked with verifying the generated questions found that 100/100 of a random sample of generated questions are aligned with the reference document and can be answered. For statement parsing, the first and second doctors found 330/330 and 329/330 of parsed statements were correctly contained in the full response, respectively. Conversely, they found 6 and 5 (out of 72 total) full responses where a single statement was not parsed, respectively.
Source verification
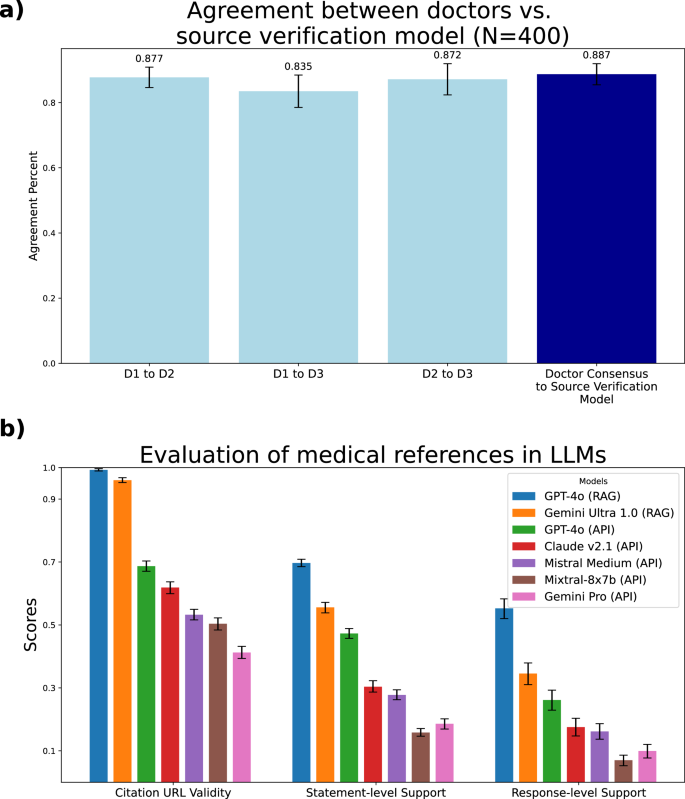
Our expert annotation of 400 statement-source pairings revealed that the Source Verification model performs as well as experts at determining whether a source supports a statement. We observed an 88.7% agreement between the Source Verification model and the doctor consensus and an 86.1% average inter-doctor agreement rate (Fig. 1a, Supplementary Table 1, Supplementary Fig., 1). We found no statistically significant difference between the doctor consensus annotations and Source Verification model annotations (p = 0.21, unpaired sample two-sided t-test).
a Agreement between the Source Verification model and doctors on the task of source verification. We asked three medical doctors (D1, D2, and D3) to determine whether pairs of statements and source texts are supported or unsupported. We found that the Source Verification model has a higher agreement with the doctor consensus than the average agreement between doctors. The 95% confidence intervals are computed with the bootstrap method and are shown in the error bars, and the total sample size is N = 400. b Evaluation of the quality of source verification in LLMs on medical queries. Each model is evaluated on three metrics. Source URL Validity measures the proportion of generated URLs that return a valid webpage. Statement-level Support measures the percentage of statements that are supported by at least one source in the same response. Response-level Support measures the percentage of responses that have all their statements supported. Full numerical results are displayed in Supplementary Table 4. The 95% confidence intervals are computed with the bootstrap method and are shown in the error bars. The sample sizes used to compute each statistic are found in Supplementary Table 8.
Evaluation of bias of GPT-4o as the backbone LLM
As an evaluation agent, Claude Sonnet 3.5 agrees with the human experts’ consensus 87.0% (83.4–90.4 95% CI) of the time, which is not statistically different than GPT-4o’s 88.7% agreement with experts (p = 0.52, paired two-sided t-test). Moreover, we observed a 90.1% (89.7–90.5 95% CI) agreement between Claude Sonnet 3.5 and GPT-4o on source verification decisions. Additionally, we performed a chi-squared test and found no statistically significant difference between using Claude or GPT-4o as the question generator or response parser (p = 0.801, paired two-sided t-test) on downstream statement support metrics (Supplementary Tables 2 and 3). These findings indicate that our evaluation pipeline is not biased towards GPT-4o and can be effectively adapted to other high-performing LLMs. We also evaluate Llama 3.1 70B on the task of citation verification and report 79.3% (75.4–83.1 95% CI) concordance with human expert consensus. As such, we find that the open-source model is not yet on-par with top proprietary models for producing expert-level citation verification.
Evaluation of source veracity in LLMs
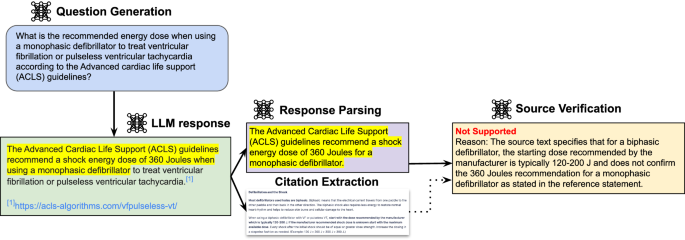
Our full results of these three metrics across seven models are found in Fig. 1b and Supplementary Table 4. We found that GPT-4o (RAG) is the highest-performing model in terms of providing citations, mainly driven by its unique ability among the models to have access to the internet via search. However, we still found that its response-level support is only 55%. We provide examples of failures from GPT-4o (RAG) in Supplementary Fig. 2, where one statement is not found due to it not being mentioned, and another is actually contradicted by a provided source. Similarly, only 34.5% of Gemini Ultra 1.0 with RAG’s responses are fully supported by the retrieved references. Additionally, the other API-endpoint models all had much lower rates across the board, largely due to the fact that they do not have access to the web. For example, GPT-4o (API), the currently best-performing LLM36, only produced valid URLs around 70% of the time. On the other end, we found that Gemini Pro’s API only produced fully supported responses about 10% of the time. Additionally, we found that open source models Llama-2-70b and Meditron-7b both were unable to consistently complete the initial task of producing citation URLs at all ( <5% for Llama and <1% for Meditron). As such, we did not include them in the main results.
As additional human expert validation, we randomly sampled 110 statement-source pairs produced by GPT-4o (RAG) that have been categorized by the Source Verification model as unsupported by any of the sources provided and had doctors assess each pair. The doctors agree with the Source Verifier 95.8% (91.8–98.7%) of the time. Among the 110 statement-source pairs provided by GPT-4o (RAG), the doctors confirmed that 105 statements are not supported by any source provided by GPT-4o (RAG). This result shows that retrieval augmentation by itself is not a silver bullet solution for making LLMs more factually accountable. In particular, while the four API-endpoint models produced sources (either valid or invalid) in >99% of responses when prompted, we find that GPT-4o (RAG) fails to produce sources in over 20% of responses, even when explicitly prompted to do so (Supplementary Fig. 2), partially contributing to its low response-level support. Overall, the average length of a cited URL source was 6905 tokens, with 99.9% of the cited URL sources falling within the context size limit for GPT-4o of 128k tokens.
Breakdown by question source
We ask whether the type of question affects the quality of sources provided by LLMs. We found that the question source significantly affected every model’s ability to produce supporting sources (detailed in Supplementary Fig. 3). For example, while the response-level support for questions from MayoClinic is close to 80% for GPT-4o (RAG), this drops precipitously to around 30% on Reddit r/AskDocs (p < 0.001 using unpaired sample t-test). Whereas questions from MayoClinic can be more directly answered from single sources, the Reddit r/AskDocs questions are more open-ended and often require pulling sources from a wide variety of domains.
Additional validation on HealthSearchQA
In addition to the real-world questions gathered from Reddit’s r/AskDocs subreddit, we also include HealthSearchQA4, a dataset of consumer health questions released by Google for the Med-PaLM paper. On a random subset of 300 questions from this dataset, we evaluated GPT-4o w/ RAG and report citation URL validity of 100%, statement-level support of 75.7% (74.0–77.2 95% CI), and response-level support of 38.4% (26.7, 49.3 95% CI). This is in line with the response-level support rate in our human-generated Reddit dataset of 31.0% (26.7, 35.8 95% CI). This provides additional support for the claim that LLMs have difficulty producing faithful citations on open-ended questions from users.
End-to-end full human evaluation
We conduct a detailed end-to-end human expert evaluation of LLM citations. In this task, a human clinician is tasked to assess a yes/no decision on whether all of the facts presented in a response are supported by the citations. This experiment evaluates GPT-4o w/ RAG on a subset of 100 questions from HealthSearchQA. The human expert finds that only 40.4% (30.7, 50.1) of the responses are fully supported by the citations provided. In comparison, on the same questions, SourceCheckup finds 42.4% (32.7, 52.2 95% CI) of responses supported. Both the human expert and SourceCheckup yield similar response-support rates on the same dataset, validating the robustness of our automated approach. Notably, both evaluation approaches support our main finding that frontier LLM with RAG do not accurately reflect sources for many medical questions.
URL analysis
We found that the URLs generated by the LLMs are predominantly from health information websites like mayoclinic.com or government health websites (e.g., nih.gov, cdc.gov) (Table 1). We also found low rates of URLs coming from paywalled or defunct web pages (Table 2). Interestingly, most sources are from US-based websites (average of 92%), with Gemini Ultra 1.0 (RAG) having the highest proportion of non-US sources (10.68%). Finally, we found that most sources are from.org or.gov domain names, indicating an origin of professional/non-profit organizations and governmental resources (Supplementary Fig. 4).
Editing model responses to improve statement relevance
We used our SourceCleanup agent to either remove or modify previously unsupported statements from GPT-4o (RAG), GPT-4o (API), and Claude v2.1 (API). On 150 unsupported statements, SourceCleanup removed 34.7% (52/150) of unsupported statements entirely. We had human experts re-evaluate the remaining 98 SourceCleanup modified statements and found that 85.7% (84/98) of the statements were now supported by the source after modification. In aggregate, the SourceCleanup agent either removes or correctly edits 90.7% (136/150) of the supported statements. We have included examples of the modifications made in Supplementary Table 5, and the entire set of modifications in our GitHub repository.
Discussion
Sourcing high-quality medical annotations can be prohibitively costly and difficult to find. While previous works have used LLMs to confirm source attribution, our work validates automated medical source verification with a panel of medical experts. Our fully automated framework allows for the rapid development of question-answering datasets while reducing the need for additional manual annotations. This capability is key, especially in the field of clinical medicine, where standard-of-care and up-to-date knowledge are constantly evolving. Additionally, our experiments with SourceCleanup also show the promise of an LLM-based approach to response editing to improve source faithfulness.
To support future research, we have structured our dataset of 58,000 statement-source pairs in a reusable format. Researchers can utilize the questions and associated sources to evaluate LLMs’ performance on source attribution across different model versions. Additionally, the statement-source pairs serve as a valuable benchmark for comparing model improvements in citation accuracy and source relevance over time, enabling longitudinal studies on the reliability of LLM-generated medical references.
Our results highlight a significant gap in the current LLMs and the desired behavior in medical settings. Regulators, clinicians, and patients alike require that model responses be trustworthy and verifiable. Central to this is that they can provide reputable sources to back their medical claims. Given that LLMs are predominantly trained on next-token prediction, it is unsurprising that “offline” models such as Mistral, Gemini-Pro, and Claude would provide hallucinated URLs or related, but incorrect, URLs as sources. We believe that if given access to the web search, these models would perform much better at producing valid URLs. To remedy this issue, models should be trained or fine-tuned directly to provide accurate source verification. RAG models show promise, as they can directly pull information from articles via search engines. However, we find that a substantial fraction of the references provided by RAG do not fully support the claims in GPT-4o (RAG) or Gemini Ultra 1.0 (RAG)‘s responses. This might be due to the LLM extrapolating the retrieved information with its pretraining knowledge or hallucination.
An important distinction of our work is that we emphasize verifying whether the LLMs’ generated statements are grounded in verifiable sources rather than directly assessing the correctness of each claim. We take this approach because the nature of whether a claim is true or false can be up to subjective interpretation—indeed, even medical experts may disagree over the degree to which a medical claim is fully factual. For example, our dataset contains a question, “What age group is most commonly affected by tennis elbow?”, which has multiple overlapping answers (e.g., 30–60 years, 30–50 years, 40–60 years). The process of ground-truthing opens up ambiguity around how to adjudicate different answers.
We find that models’ responses to Reddit questions are on average, longer than those from MayoClinic, given the more open-ended nature and tendency for users to ask about multiple related topics. Additionally, the model is more speculative in its responses to Reddit questions, often providing more disclaimers and potential answers. These factors lead to both the statement-level response-level support being lower on Reddit questions compared to the MayoClinic. In general, we notice that for direct questions with straightforward answers, models are much stronger at providing relevant citations. When models are asked to speculate or provide multiple answers, they tend to produce responses that deviate further from the provided citations.
Under Section 230 of the Communications Decency Act, websites like Twitter or WebMD are not regulated by the US FDA, since they simply act as an intermediary for, rather than the author of, medical information37. However, it is unclear if this existing legal protection is likely to apply to LLMs since they can extrapolate and hallucinate new information. Additionally, the existing regulatory framework for AI software medical devices may also not apply to LLMs, as they do not have constrained, deterministic outputs38. Thus, assessing the degree to which LLMs reliably convey existing, trustworthy medical knowledge is important for informing future regulatory frameworks regarding medical LLMs.
In our breakdown of source verification by question source, we find that models perform significantly worse on questions sourced from Reddit r/AskDocs versus MayoClinic. This is significant, as questions from Reddit are user-generated, whereas MayoClinic is vetted by medical professionals. One potential reason for this divergence is that user-generated questions tend to reflect a more diverse distribution of topics and more variable reading levels than medical reference sites, which tend to use precise medical terminology. Another hypothesis is that user-generated questions may contain erroneous premises for which LLMs have the propensity to affirm, known as contra-factual bias39. In this same vein, we also found that the cited URLs are from US-based sources over 90% of the time, which may potentially reflect American patient-centric medical evidence and standard of care. It is important thus for LLMs to adequately perform source verification to serve a wide range of users—both laypersons and medical professionals, as well as sources that represent their intended demographic.
Measuring source verification in models is also not intended to be considered in isolation. For example, one could trivially perform perfectly by quoting verbatim from a set of known sources (e.g., Google search). Instead, this benchmark should be used with other quality-based evaluation metrics to highlight inherent trade-offs in LLMs when they extrapolate information. To this end, we are releasing all of our curated data and expert annotations as a community resource.
Our approach also has several limitations that motivate follow-up studies. First, our automated pipeline can accrue errors in the question generation, statement parsing, citation extraction, and source verification stages. While we have performed spot checks on each component, these errors are non-zero and can lead to small fluctuations in the final reported results. Second, the task of source verification can be ambiguous, as shown by the lack of full agreement among the three medical doctors in our study. As such, while we believe our final results represent an accurate scale of the relevance of medical queries, individual data points may be more noisy and prone to interpretation. Third, our usage of a 1-1 mapping between statements and sources has a tradeoff such that we do count cases where a statement can be supported by aggregating multiple sources together. However, as an experiment to evaluate the extent to which this effect is occurring, we analyzed all statements judged as unsupported from GPT-4o (RAG) and re-ran the analysis with the sources merged for each statement. We find that 95.1% of the statements previously unsupported are still not supported after merging the sources. Our URL analysis findings report a high rate of US-based websites, which could be in part due to the fact that our questions are largely drawn from US-based sources (e.g., MayoClinic). Finally, our URL content extraction module is prone to a small rate of 404 errors on websites that are otherwise accessible to individuals in websites that prevent repeated requests from the same user agents. Additionally, the ability to access scientific texts behind paywalls depends on each researcher’s access, meaning that there may be discrepancies in URL validity when run across institutions.
We believe that going forward, source verification is key to ensuring that doctors have accurate and up-to-date information to inform their clinical decision-making and provide a legal basis for LLMs to be used in the clinic. Indeed, accurate source verification extends beyond the medical domain and has apt applications in other fields like law (e.g., case law) and journalism (e.g., fact-checking).
Methods
This study was conducted in strict accordance with all applicable ethical standards, guidelines, and regulations governing research practices.
LLM evaluations
Our analysis focuses on evaluating the following top-performing LLMs: GPT-4o (RAG), GPT-4o (API), Claude v2.1 (API), Mistral Medium (API), Gemini Ultra 1.0 (RAG), and Gemini Pro (API). These models were chosen as they represent current leading LLMs3,36,40,41 as of February 2024. Additionally, we consider the following open-source models: Mixtral-8x7b (API), Llama-2-70b (API), and Meditron-7b, where Meditron-7b is a medical-domain open-source model. We use gpt-4o-2024-05-13 as the GPT-4o API endpoint, while Gemini Ultra 1.0 (RAG) was evaluated on 3/28/24. All other model APIs were queried for responses on 1/20/24. The parenthetical (RAG) refers to the model’s web browsing capability powered by web search. When not labeled with (RAG), GPT-4o refers to the standard API endpoint used in this study, without web browsing capability.
SourceCheckup evaluation framework
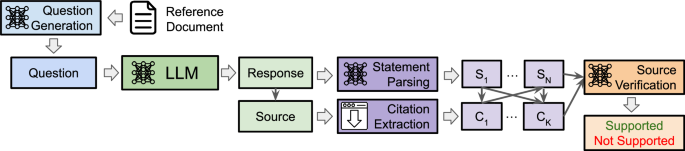
Our proposed pipeline consists of four modules: (1) Question Generation, (2) LLM Question Answering (3) Statement and URL Source Parsing, and (4) Source Verification. A schematic of this pipeline is found in Fig. 2, and an example is shown in Fig. 3. The prompts used for each of the following sections are detailed in Supplementary Table 6.
To start, GPT-4o generates a question based on a given medical reference text. Each evaluated LLM produces a response based on this question, which includes the response text along with any URL sources. The LLM response is parsed for individual medical statements while the URL sources are downloaded. Finally, the Source Verification model is asked to determine whether a given medical statement is supported by the source text and to provide a reason for the decision.
A question is generated based on the contents of a medical reference text. The question is posed to an LLM, and the response is parsed into statements and sources. Each statement-source pair is automatically scored by the Source Verification model as supported (i.e., the source contains evidence to support the statement) or not supported.
Question generation
First, we collected 400 real-world medical queries from Reddit’s r/AskDocs, a subreddit for patients to ask medical professionals that has over 600 K members. These questions are typically presented as short cases with relevant symptoms provided by users. Additionally, given that medical question datasets such as PubMedQA42 and MedQA43 consist of fixed question sets susceptible to memorization, we propose a question generation framework to create medical questions that reflect real-world clinical question/answering. A reference text was given to GPT-4o with a prompt to produce a question based on the content of the text. In this study, we select reference texts from MayoClinic, which provides patient-facing fact pages on common medical queries. MayoClinic content allows us to generate text comprehension-based questions, which may differ in style and tone from natural user queries. None of our reference documents were taken from private datasets containing protected health information. We used GPT-4o to generate a question from each of the 400 reference documents from MayoClinic. We then combined these 400 generated questions with 400 real-world queries from r/AskDocs to produce our full set of 800 questions. Finally, we posed each question to each of the seven LLMs. We include several examples of questions in Supplementary Table 7.
LLM question answering
We queried each LLM to provide a short response to the question, along with a structured list of sources that support the response. The prompt used for querying LLMs can be found in Supplementary Table 6. To gather responses from the GPT-4o (RAG) model with web browsing capabilities, we found that the standard prompt was unable to trigger the web search RAG capabilities, so we provided a modified version of the prompt that explicitly asks the model to use Bing Search. In a minority of cases, the model did not return a response or returned an incomplete response. In this event, we provided the LLM an additional try before considering the response invalid.
Statement parsing
To break up the response into individually verifiable statements, we used GPT-4o to parse the LLM responses. We define a “statement” to be an independently verifiable part of a model’s response. For example, the response “The proportion of HFE C282Y homozygotes with documented iron overload-related disease is 28.4% for men and 1.2% for women” is broken into “The proportion of HFE C282Y homozygotes with documented iron overload-related disease is 28.4% for men” and “The proportion of HFE C282Y homozygotes with documented iron overload-related disease is 1.2% for women”. Certain responses did not return any parsed medical statements, largely due to the nature of the question asked. For example, the model response “Could you please provide the document or specify the details of the treatment options and durations for acute bacterial rhinosinusitis mentioned in it?” does not return any parsed medical statements. Full details of the number of parsed statements, along with source counts, are found in Supplementary Table 8. Additionally, more details on the human instructions used in our validation are in Supplementary Table 9. In general, we find that GPT-4o (API) and Claude v2.1 could consistently follow the instructions’ JSON formatting, whereas other models had varying rates of success. In cases where the model fails to provide a structured source list, we extracted and removed all URLs using regular expression matching from the original text and treated them as the sources provided.
URL source parsing
For each URL source provided in the response, we downloaded the source content of the URL. We only kept websites that returned a 200 status code, meaning the content can be returned. A small percentage ( < 1%) of cases also included websites that cannot be accessed through our pipeline. We downloaded PDF documents locally before extracting their text using a PDF-to-text converter. After the source content was extracted, we applied a pattern-matching expression to strip code tags, leaving only the plain text. Finally, we excluded source contents that exceed the 128 K maximum token length of GPT-4o. This accounted for approximately 0.1% of all downloaded URLs.
Source verification
We considered a statement to be supported if it can be attributed to at least one source provided by the LLM. While not all sources are usually intended to support each statement, we found the task of determining each LLM’s intended statement-source pairings to be difficult to determine. For example, some LLMs provided a footnote after each statement, whereas others provided a list of links at the end of a paragraph. As such, we opted to simply consider all pairs of statements and sources in our evaluation. Additionally, we individually evaluate each source’s attribution to each statement individually rather than combining all given sources together. This was done so for two reasons: (1) a 1-1 mapping for sources and statements allows us to report a “precision” metric (what % of sources do not support any statement in the response). This measures each model’s ability to intentionally attribute their statements to sources rather than generate a long laundry list of citations from a generic search result page, and (2) we found the task of asking doctors to verify if the information has been properly integrated across multiple documents to be more complex and ambiguous (eg. What is the proper way to perform meta-analyses across studies, what if articles contradict one another, how does one weigh one article’s credibility vs another, etc.).
Given a list of statements and sources, each possible pair was checked for whether the source contained the relevant information necessary to support the statement. For instance, given \(M\) statements and \(N\) sources, each of the \(M\) statements was checked against each of the \(N\) sources for a total of \({MxN}\) pairs. For each pair, we prompted GPT-4o with the statement and source content and asked it to score the pair. If a statement was supported by at least one source, it was considered “supported”; otherwise, it was considered “not supported”. To disambiguate the use of GPT-4o for source verification as well as evaluation, we refer to this task as the “Source Verification model” and the evaluated model by the full name (i.e., GPT-4o (RAG) or GPT-4o (API)).
Rashkin et al. propose23 and formalize a framework called Attributable to Identifiable Sources, which defines the AIS score of a given language model’s response, \(y\), supported by evidence Aas 1 if a human reviewer would agree that “\(y\) is true, given \(A\)” or 0 if not. Gao et al. extends24 this to measure the average sentence-level AIS score:
$${Att}{r}_{{AIS}}(y,\,A)={av}{g}_{s \, \in \, y}{AIS}(s,\,A)$$
which is the percentage of statements within a response that is fully supported by \(A\). We extend these two definitions of statement-level and response-level AIS scores to statement-level and response-level support below.
We report three metrics to evaluate each model’s source verification capabilities:
Source URL validity
Given all the source URLs produced by the model, what percent are valid? We define a valid URL as one that produces a 200 status code when requested and returns valid text (non-empty response).
$${Source\; URL\; Validity}=\frac{{{{\rm{\#}}}}{URLs\; with\; status\; code} \, 200}{{Total\; Number\; of\; URLs}}$$
Statement-level support
What percent of medically relevant statements produced by the model can be supported by at least one source? For each statement parsed from the responses, we checked it against all sources produced by the model response. A statement was considered supported if at least one of those sources was found to contain supporting text.
$${Statement\; Level\; Support}=\frac{{Statements\; Supported\; by}\ge \!1 \, {Source}}{{Total\; Number\; of\; Statements}}$$
We note that this metric does not penalize LLM responses for producing many irrelevant sources. To this end, we also report the percent of URLs that are not used in supporting any statement, found in Supplementary Table 10.
Response-level support
What percent of responses have all their statements supported? For each response, we checked whether that response contained all supported statements.
$${Response\; Level\; Support}=\frac{{Responses}\, w/{All\; Statements\; Supported}}{{Total\; Number\; of\; Resonses}}$$
Expert validation of GPT-4o automated tasks
Each of the three GPT-4o automated tasks (Question Generation, Response Parsing, and Source Verification) was validated against the annotations of US-licensed practicing medical doctors.
Question generation and response parser
To validate the performance of GPT-4o on the task of generating questions from reference medical documents, we asked two medical doctors to spot-check 100 pairs of documents and questions for relevance and logical integrity. To validate GPT-4o’s performance on parsing medical statements from free-text responses, we also asked two medical doctors to analyze a sample of 330 statements from 72 question/response pairs to check (1) if all the statements are found within the original response, and (2) if any statements are missing from the list of parsed statements.
Source verification
A subset of the statement and sources (N = 400) was selected from model responses from GPT-4o (RAG), GPT-4o (API), and Claude v2.1 (API). Three medical doctors independently scored whether the LLM-generated source verification decision correctly identified a statement as supported or not supported by the provided source. They also optionally provided a reason justifying their decision. We then calculated the majority consensus of the doctors and reported the percent agreement among each doctor, the doctor consensus, and the LLM-generated decision.
Potential bias of GPT-4o as an evaluator, parser, and question generator
We aim to evaluate whether our use of GPT-4o as the backbone of our pipeline introduces downstream biases in the support rates of models. To do so, we replicated our entire pipeline using Claude Sonnet 3.5, a leading LLM with comparable performance to GPT-4o. Using the same set of original documents from MayoClinic, we re-generate questions, parse model responses, and perform source verification with Claude Sonnet 3.5. Additionally, we benchmark Llama 3.1 70B on the task of citation verification to evaluate the capabilities of an open-source model. Finally, we compare the statement support rates between statement-source pairs which have been produced by replacing GPT-4o with Claude Sonnet 3.5 in each part of our pipeline.
Improve statement relevance to sources
Unsupported statements often only partially deviate from their source. Given this, we explore how effectively LLMs can revise these unsupported statements to make them fully supported by the original source. To address this, we developed an LLM agent called “SourceCleanup.” This agent uses GPT-4o as the backbone model and takes a single statement and its corresponding source as input and returns a modified, fully supported version of the statement. The prompt used for SourceCleanup is detailed in Supplementary Table 6.
URL analysis
We computed several key statistics from the total set of URLs cited per model. First, we determined which domain names contain content that is hidden behind a paywall or subscription model. Second, of the URLs that were deemed invalid due to a 404 error or similar “page not found” response, we assessed how many URLs were previously valid but are now outdated. To approximate this, we used the Internet Archive Wayback Machine API, which stores archived URLs. Second, we reported the top five domain names cited by each model.
Finally, we analyzed the origin of URLs in two ways. First, we determined which domain names are US-based vs non-US-based by performing a whois lookup and looking for a valid country. If no country is returned, we default to count domains with TLDs contained in.com,.org,.gov,.edu,.info,.net as US-based, and non-US-based if not.
Statistics and reproducibility
This study utilized an automated, agent-based evaluation framework, SourceCheckup, to systematically assess the relevance and supportiveness of medical references cited by large language models (LLMs). No statistical method was used to predetermine sample sizes, and no data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. Statistical analyses included calculation of percent agreement between automated Source Verification models and human expert consensus, along with comparisons using paired and unpaired two-sided t-tests and chi-squared tests, as appropriate. Reproducibility was confirmed through independent validation by US-licensed medical experts, and code and data for reproducibility are publicly available in our GitHub repository (https://github.com/kevinwu23/SourceCheckup).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The questions, responses, parsed statements, fact-citation pairs, and expert annotations generated in this study have been deposited in a Google Drive accessible through the following open-access link: https://drive.google.com/drive/folders/1a-i974g3XzLCtZLpTLBbqAwK0olpd5JY?usp=drive_link. This open-access data is available in perpetuity exclusively for non-commercial research purposes. The raw web page files from citations are not included in our data due to restrictions from some content providers; however, researchers can access the URLs themselves if they have appropriate permissions. For any inquiries regarding access, please contact [email protected] (please allow 3-5 days response time).
Code availability
The code to implement SourceCheckup’s pipeline, as well as the prompts used in each of the agent modules, can be found at https://github.com/kevinwu23/SourceCheckup. DOI identifier: https://doi.org/10.5281/zenodo.15015209.
References
Gilbert, S., Harvey, H., Melvin, T., Vollebregt, E. & Wicks, P. Large language model AI chatbots require approval as medical devices. Nat. Med. 29, 2396–2398 (2023).
Brin, D. et al. Comparing ChatGPT and GPT-4 performance in USMLE soft skill assessments. Sci. Rep. 13, 16492 (2023).
Singhal, K. et al. Toward expert-level medical question answering with large language models. Nat. Med. 31, 1–8 (2025).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Ingram, D. ChatGPT used by mental health tech app in AI experiment with users. NBC News (2023).
Maples, B., Cerit, M., Vishwanath, A. & Pea, R. Loneliness and suicide mitigation for students using GPT3-enabled chatbots. NPJ Ment. Health Res. 3, 1–6 (2024).
Holohan, M. A boy saw 17 doctors over 3 years for chronic pain. ChatGPT found the diagnosis. TODAY (2023).
Temsah, M.-H. et al. ChatGPT and the future of digital health: a study on healthcare workers’ perceptions and expectations. Healthcare. 11, 1812 (2023).
Tangadulrat, P., Sono, S. & Tangtrakulwanich, B. Using ChatGPT for clinical practice and medical education: cross-sectional survey of medical students’ and physicians’ perceptions. JMIR Med Educ. 9, e50658 (2023).
Pal, A., Umapathi, L. K. & Sankarasubbu, M. Med-HALT: medical domain hallucination test for large language models. Proc. 27th Conf. Comput. Nat. Lang. Learn. (CoNLL), 314–334 (2023).
Sun, L. et al. TrustLLM: trustworthiness in large language models. Proc. 41st Int. Conf. Mach. Learn. (2024).
Ahmad, M. A., Yaramis, I. & Roy, T. D. Creating trustworthy LLMs: dealing with hallucinations in healthcare AI. arXiv 2311.01463 Available at: https://arxiv.org/abs/2311.01463 (2023).
Dash, D. et al. Evaluation of GPT-3.5 and GPT-4 for supporting real-world information needs in healthcare delivery. arXiv 2304.13714 Available at: https://arxiv.org/abs/2304.13714 (2023).
Daws, R. Medical chatbot using OpenAI’s GPT-3 told a fake patient to kill themselves. AI News Available at: https://www.artificialintelligence-news.com/news/medical-chatbot-openai-gpt3-patient-kill-themselves/ (2020).
Nastasi, A.J., Courtright, K.R., Halpern, S.D. et al. A vignette-based evaluation of ChatGPT’s ability to provide appropriate and equitable medical advice across care contexts. Sci. Rep. 13, 17885 (2023).
Zawiah, M. et al. ChatGPT and clinical training: perception, concerns, and practice of Pharm-D students. J. Multidiscip. Heal 16, 4099–4110 (2023).
Abouammoh, N. et al. Perceptions and earliest experiences of medical students and faculty with ChatGPT in medical education: qualitative study. JMIR Med. Educ. 11, e63400 (2025).
Jansz, J. & Sadelski, P. T. Large Language Models in Medicine: The potential to reduce workloads, leverage the EMR for better communication & more. Rheumatologist (2023).
Hacker, P., Engel, A. & Mauer, M. Regulating ChatGPT and other Large Generative AI Models. in Proceedings of the 2023 ACM Conference on Fairness, Accountability, and Transparency 1112–1123 (Association for Computing Machinery, 2023).
Baumann, J. ChatGPT Poses New Regulatory Questions for FDA, Medical Industry. Available at: https://news.bloomberglaw.com/health-law-and-business/chatgpt-poses-new-regulatory-questions-for-fda-medical-industry (2023).
Taylor, N. P. FDA calls for ‘nimble’ regulation of ChatGPT-like models to avoid being ‘swept up quickly’ by tech. Available at: https://www.medtechdive.com/news/fda-calls-for-nimble-regulation-of-chatgpt-like-models-to-avoid-being-sw/649756/ (2023).
Li, X., Cao, Y., Pan, L., Ma, Y. & Sun, A. Towards verifiable generation: a benchmark for knowledge-aware language model attribution. Findings Assoc. Comput. Linguist.: ACL 2024, 493–516 (2024).
Rashkin, H. et al. Measuring attribution in natural language generation models. Comput. Linguist. 49, 777–840 (2023).
Gao, T., Yen, H., Yu, J. & Chen, D. Enabling large language models to generate text with citations. Proc. 2023 Conf. Empir. Methods Nat. Lang. Process., 6465–6488 (2023).
Nakano, R. et al. WebGPT: Browser-assisted question-answering with human feedback. Available at: https://arxiv.org/abs/2112.09332 (2021).
Malaviya, C. et al. ExpertQA: expert-curated questions and attributed answers. Proc. 2024 Conf. North Am. Chapter Assoc. Comput. Linguist.: Hum. Lang. Technol., 3025–3045 (2024).
Proposed Taxonomy for Gender Bias in Text - ACL Anthology. Httpsaclanthologyorg › Httpsaclanthologyorg ›.
Kamalloo, E., Jafari, A., Zhang, X., Thakur, N. & Lin, J. HAGRID: A Human-LLM Collaborative Dataset for Generative Information-Seeking with Attribution. Available at: https://arxiv.org/abs/2307.16883 (2023).
Chen, H.-T., Xu, F., Arora, S. & Choi, E. Understanding Retrieval Augmentation for Long-Form Question Answering. (2023). Proc. 2024 Conf. Lang. Model. (COLM), (2024).
Bohnet, B. et al. Attributed Question Answering: Evaluation and Modeling for Attributed Large Language Models. Available at: https://arxiv.org/abs/2212.08037 (2022).
Menick, J. et al. Teaching language models to support answers with verified quotes. Available at: https://arxiv.org/abs/2203.11147 (2022).
Min, S. et al. FActScore: fine-grained atomic evaluation of factual precision in long form text generation. Proc. 2023 Conf. Empir. Methods Nat. Lang. Process., 12076–12100 (2023).
Yue, X. et al. Automatic evaluation of attribution by large language models. Findings Assoc. Comput. Linguist.: EMNLP 2023, 4615–4635 (2023).
Zuccon, G., Koopman, B. & Shaik, R. ChatGPT Hallucinates when Attributing Answers. Available at: https://arxiv.org/abs/2309.09401 (2023).
Liu, N., Zhang, T. & Liang, P. Evaluating verifiability in generative search engines. Findings Assoc. Comput. Linguist.: EMNLP 2023, 7001–7025 (2023).
Tatsu. AlpacaEval Leaderboard. (2024).
Haupt, C. E. & Marks, M. AI-generated medical advice-GPT and beyond. JAMA 329, 1349–1350 (2023).
Gottlieb, S. & Silvis, L. How to safely integrate large language models into health care. JAMA Health Forum 4, e233909 (2023).
Dahl, M., Magesh, V., Suzgun, M. & Ho, D. E. Large legal fictions: profiling legal hallucinations in large language models. J. Leg. Anal. 16, 64–93 (2024).
Eriksen Alexander, V., Sören, M. öller & Jesper, Ryg Use of GPT-4 to diagnose complex clinical cases. NEJM AI 1, AIp2300031 (2023).
Strong, E. et al. Chatbot vs medical student performance on free-response clinical reasoning examinations. JAMA Intern. Med. 183, 1028–1030 (2023).
Jin, D. et al. What disease does this patient have? A large-scale open domain question answering dataset from medical exams. Appl. Sci. 11, 6421 (2021).
Jin, Q., Dhingra, B., Liu, Z., Cohen, W. W. & Lu, X. PubMedQA: A Dataset for Biomedical Research Question Answering. (2019).
Acknowledgements
We thank Josiah Aklilu and Min Woo Sun for their helpful feedback on our paper.
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yifan Peng, Aixin Sun, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, K., Wu, E., Wei, K. et al. An automated framework for assessing how well LLMs cite relevant medical references. Nat Commun 16, 3615 (2025). https://doi.org/10.1038/s41467-025-58551-6
Received: 30 September 2024
Accepted: 26 March 2025
Published: 16 April 2025
DOI: https://doi.org/10.1038/s41467-025-58551-6
.png)