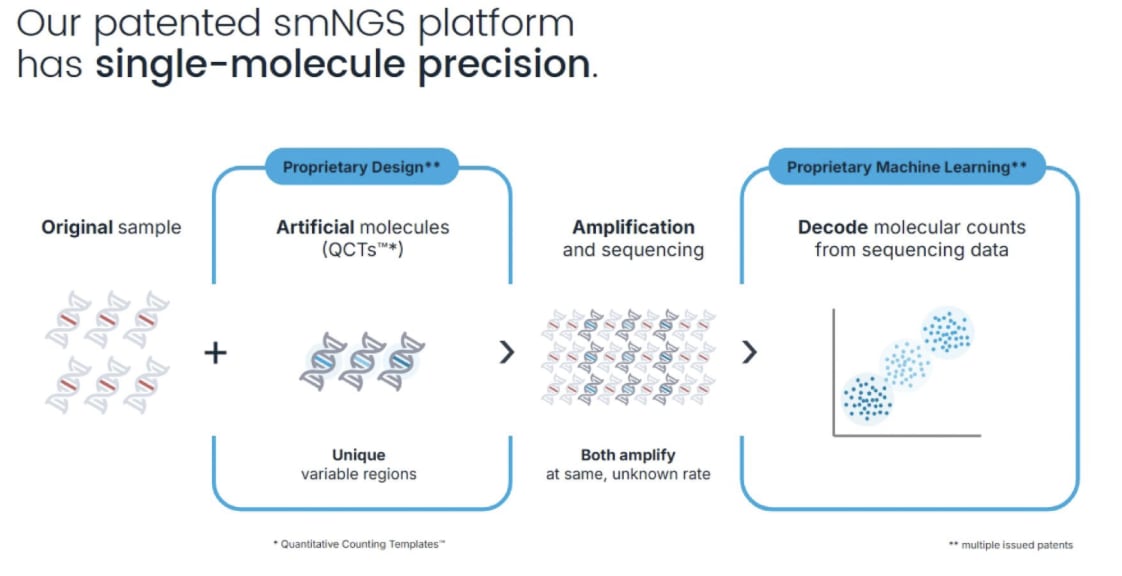
BillionToOne is filing for IPO. They have an exciting “single molecule NGS” solution. If that was really single molecule that would be very exciting! After all we have two single molecule sequencers on the market (PacBio and ONT). So if BillionToOne was deriving significant value ($150M+ revenue) from single molecule sequencing, this would be quite exciting.
But BillionToOne isn’t “single molecule sequencing” in the way anyone else uses the term “single molecule sequencing”, that is to say “sequencing single molecules”. This is… spike in controls!
BillionToOne don’t disclose the backend sequencing tech, so it could be PacBio or ONT. But for a counting application like this using short reads, it seems far more likely they are using Illumina or similar short read sequencers.
So, is every NGS platform released since the Solexa Genome Analyzer in 2006 a single molecule sequencer? If you think so BillionToOne is “single molecule NGS”. But this seems like an odd way to use the term…
What does BillionToOne use this spike-in calibration technology for? NIPT:
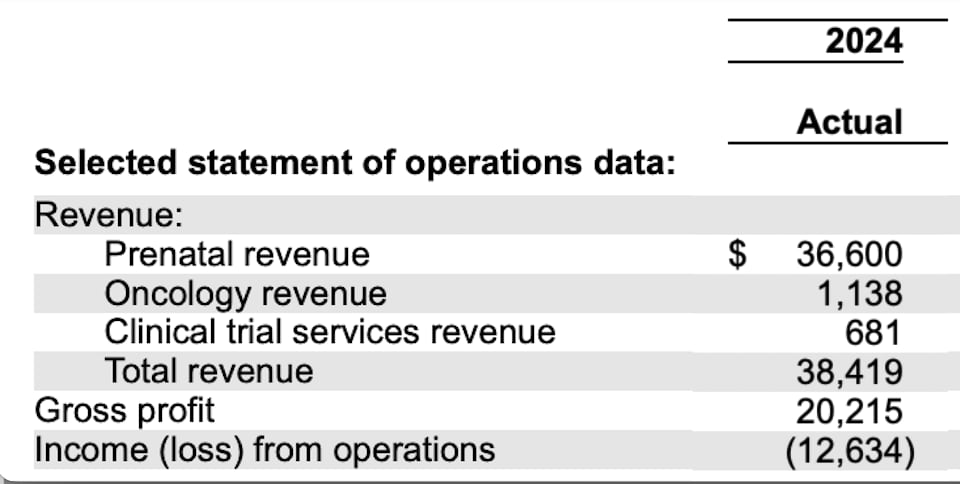
They run about 500K tests a year (running at a loss).
The main sell seems to be an expanded panel looking at point mutations causing “SCD, alpha thalassemia, beta-thalassemia, CF, and SMA”.
I personally would be surprised if their spike-in calibration was required to enable this, other approaches seem to work. If some kind of calibration is required, I would be surprised if the IP would hold up sufficiently to exclude other players from using similar approaches.
It seems like Natera are also moving into this space. So I’d expect BillionToOne to see increasing competition here, and I’m doubtful that Natera need BillionToOne’s IP to enable this when they launch.
That said, BillionToOne appears to have a reasonable foothold in this market. So it will be interesting to see how it all plays out.
Was this useful? Please subscribe. Even free subscription help grow the substack.
.png)