I’ve just shipped StringZilla v4, the first CUDA-capable release of my SIMD-first string processing library. Which in English means that it is now fast not only on CPUs, but also on GPUs!
- I’ve wanted to add ROCm-acceleration for AMD GPUs 🤦♂️
- I’ve wanted to include a parallel multi-pattern search algorithm 🤦♂️
- I’ve wanted to publish it back in December 2024, not September 2025 🤦♂️
So not everything went to plan, but “StringZilla 4 CUDA” is finally here, bringing 500+ GigaCUPS of edit-distance calculations in a free pip install-able package, and a few more tricks up its sleeve, aimed at large-scale Information Retrieval, Databases and Datalake systems, as well as Bioinformatics workloads. So in this post, we’ll cover some of the most interesting parts of this release, including:
- Fast evaluation of dynamic-programming algorithms on GPUs,
- High CPU-port-level parallelism in SIMD code, and
- Fingerprinting biological sequences with 52-bit integers.
Background & Inspiration
Historically, StringZilla started as conference talk material in the late 2010s, showcasing the power of AVX-512 and the intricacies of vectorizing non-data-parallel workloads (… pretty much the opposite of my SimSIMD). Over the years, it expanded from a few substring search kernels into a beast competing with GLibC for the fastest memcpy (yes, I know it’s a popular claim). It later added support for little- & big-endian platforms; several generations of AVX-512 on x86, Arm NEON, SVE, and SVE2 extensions; dynamic dispatch; and first-party bindings for Python, Rust, JavaScript, and even Swift, translating the underlying C99 implementation.
Now, StringZilla v4 adds even more functionality:
- Yet another non-cryptographic hash function and string PRNGs on AES and other port-parallel SIMD instructions.
- New intersection and sorting algorithms for extensive collections of strings standard in DBMS JOINs and ORDER BYs.
- GPU- and CPU-accelerated string similarity kernels, covering Levenshtein distances, Needleman-Wunsch and Smith-Waterman scores with Gotoh’s extensions for “affine gaps”, needed in bioinformatics.
- GPU- and CPU-accelerated fingerprinting MinHashing kernels for large-scale Information Retrieval and deduplication.
Depending on the chosen implementation and input data, these may be an order of magnitude faster than what you are using today. Not all of the new kernels are State-of-the-Art, as I partially demonstrate in my refactored StringWa.rs benchmarks repository — they’re solving a different problem: providing a reliable, fast, and easy-to-use baseline for large-scale workloads.
Traditional String Similarity Measures
Dynamic Programming and Levenshtein Evaluation Order
If we look at the classic description of the Levenshtein distance computing methodology, it suggests incrementally populating an $N + 1$ by $M + 1$ matrix, where $N$ and $M$ are the lengths of the two strings being compared. The order in which we fill this matrix is crucial for the algorithm’s efficiency and correctness:
$$ L_{i,j}=\min\left[ \begin{array}{l} L_{i-1,j}+1, \ L_{i,j-1}+1, \ L_{i-1,j-1}+[x_i \ne y_j] \end{array} \right] $$
$$ L_{0,j}=j, L_{i,0}=i $$
Here, $L_{i,j}$ represents the edit distance between the first $i$ characters of string $x$ and the first $j$ characters of string $y$. The three terms in the minimum represent deletion, insertion, and substitution costs respectively, where $[x_i \ne y_j]$ equals 1 if characters differ and 0 if they match.
If we look up the Wikipedia article or the absolute majority of code-snippets they describe the Wagner-Fischer algorithm, filling that matrix top-to-bottom, left-to-right. More memory-efficient implementations suggest storing only 2 rows of the matrix at any time, significantly reducing the space complexity from $O(NM)$ to $O(\min(N, M))$. That, however, does nothing to remove the sequential data dependency in the lower row - we can’t process $L_{i,j}$ without having computed all $L_{i,j-1}$.
The smarter way, when parallelizing such algorithms is to look into dependency chains that break vectorization potential. In the case of such string similarity measures, including Levenshtein distance, it’s simple - evaluate diagonals instead of rows! We’ll store 3 diagonals instead of the 2 rows, and each consecutive diagonal will be computed from the previous two. Substitution costs will come from the sooner diagonal, while insertion and deletion costs will come from the later diagonal.
| Row-by-Row Algorithm Computing row 4: ∅ A B C D E ∅ 0 1 2 3 4 5 P 1 ░ ░ ░ ░ ░ Q 2 ■ ■ ■ ■ ■ R 3 ■ ■ □ → . S 4 . . . . . T 5 . . . . . | Anti-Diagonal Algorithm Computing diagonal 5: ∅ A B C D E ∅ 0 1 2 3 4 5 P 1 ░ ░ □ □ ■ Q 2 ░ □ □ ■ ↘ R 3 □ □ ■ ↘ . S 4 □ ■ ↘ . . T 5 ■ ↘ . . . |
| Legend: 0,1,2,3... = initialization constants ░ = cells processed and forgotten ■ = stored cells □ = computing in parallel → ↘ = movement direction . = cells to compute later | |
Did it help? Performance of such algorithms is measured in Cell Updates Per Second, or CUPS. Comparing ≅ 1000-byte strings resulted in:
- rapidfuzz::levenshtein: 14'316 MCUPS on an Intel Sapphire Rapids core,
- stringzillas::LevenshteinDistances: 13'084 MCUPS on the same 1 core,
- stringzillas::LevenshteinDistances: 624'730 MCUPS on an Nvidia H100 GPU.
NLTK, one of Pythons’ historically most used libraries with over 1 Billion downloads, yields around ≅ 2 MCUPS. To my great surprise, cudf wrapping the nvtext::levenshtein implementation from Nvidia’s RAPIDS ecosystem, seemingly achieves only ≅ 1'950 MCUPS on the same H100 GPU. To be fair, cudf is not exactly a string-similarity library, and there is a separate commercial offering from Nvidia called Clara Parabricks that probably does better.
Substitution Costs and Affine Gap Penalties
In Bioinformatics, Levenshtein distance is used to compare DNA strings. Character insertions and deletions in string pairs signal physical breaks in really long molecules. The presence of those gaps is a bigger factor than the inferred length of the gap, so those are scored differently - using “affine gap penalties”. It’s not enough to store 1 matrix anymore, we need to store 3 matrices, including 2 new ones to differentiate gap extension and openings.
Similarly, Needleman-Wunsch score generalizes the Levenshtein distance to handle variable substitution costs. That’s handy when dealing with protein sequences. Those are represented as strings over a much smaller alphabet of 20 amino acids: ACDEFGHIKLMNPQRSTVWY. So one needs to store a 20x20 substitution matrix, or a larger one if we want to include ambiguous amino acids.
On the bright side, StringZilla already uses Nvidia’s DP4A and DPX instructions for SIMD processing of small integers in such dynamic programming workloads. On the other side, StringZilla’s current implementation mistakenly uses constant memory to store the substitution matrix, which is a bottleneck and will be improved in the future. Still, the results are not too bad compared to other pip install-able solutions. On ≅ 1000-long amino-acid sequences, we get:
- biopython achieving ≅ 303 MCUPS,
- stringzillas-cpus achieving ≅ 276 MCUPS,
- stringzillas-cuda achieving ≅ 10'098 MCUPS.
Two More Hash Functions?!
Design Goals
There are a lot of hash functions around! Some of them are really-really good!
- The XXH3 by Yann Collet, for example, is a remarkable production-ready codebase in which even very experienced developers may find something new.
- MurMurHash is a classic, released by Austin Appleby in 2008 together with the SMHasher framework.
- There is also a very well crafted ahash package for Rust by Tom Kaitchuck, popularizing AES instructions, also previously suggested in AquaHash by J. Andrew Rogers.
Overall, I wanted a hash function that:
- Is fast for both short (velocity) and long strings (throughput).
- Supports incremental (streaming) hashing, when the data arrives in chunks.
- Supports custom seeds for hashes and have it affecting every bit of the output.
- Provides dynamic-dispatch for different architectures to simplify deployment.
- Uses not just AVX2 & NEON SIMD, but also masked AVX-512 & predicated SVE2.
- Documents its logic and produces the same output across different platforms.
- Outputs 64-bit or 128-bit hashes and passes the SMHasher --extra tests.
AES and Port-Parallelism Recipe
Using AES instructions for hashing is not a new idea, but it makes a lot of sense. Just look at the amount of signal mixing that happens in a single round of AES:
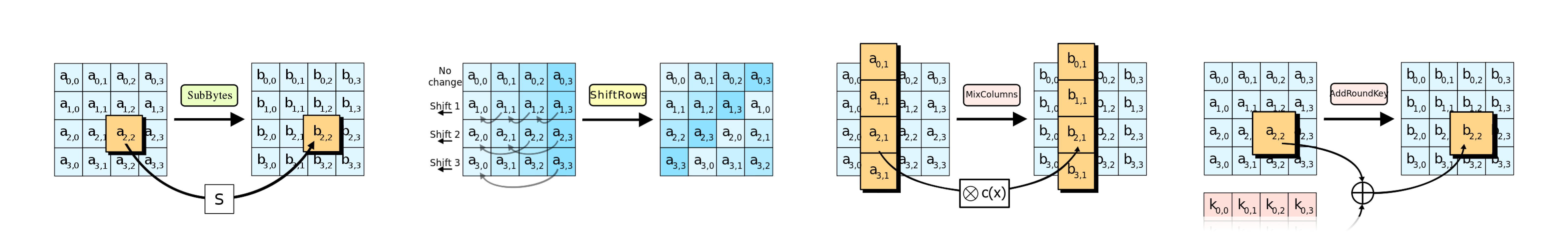
In every iteration, AES performs four operations on a $4*4=16$ byte-matrix:
- SubBytes - a non-linear substitution step where each byte is mapped to another.
- ShiftRows – a transposition where the last three rows are shifted cyclically.
- MixColumns – a linear mixing operation that operates on the columns of the state.
- AddRoundKey – a simple XOR operation with a round key.
In the second step, we mix bytes between consecutive 32-bit words of the state. In the third step, we mix bytes within a single 32-bit word. And the rest of the logic applies to the entire 128-bit matrix. Great instructions for mixing, but not the cheapest ones:
- VAESENC (ZMM, ZMM, ZMM) and VAESDEC (ZMM, ZMM, ZMM):
- on Intel Ice Lake: 5 cycles on port 0.
- On AMD Zen4: 4 cycles on ports 0 or 1.
As it often happens with integer workloads on Intel, a SIMD instruction is always routed to just one execution port, often port 0 or 5. AES routes to 0, so to mix more data in parallel we need to combine it with cheap instructions mapping to other ports, like:
- VPSHUFB_Z (ZMM, K, ZMM, ZMM)
- on Intel Ice Lake: 3 cycles on port 5.
- On AMD Zen4: 2 cycles on ports 1 or 2.
- VPADDQ (ZMM, ZMM, ZMM):
- on Intel Ice Lake: 1 cycle on ports 0 or 5.
- On AMD Zen4: 1 cycle on ports 0, 1, 2, 3.
That’s a solid recipe, and is ideologically close to ahash, but with a few twists:
- A larger state and a larger block size is used for inputs longer than 64 bytes, benefiting from wider registers on current CPUs. Like many other hash functions, the state is initialized with the seed and a set of Pi constants. Unlike others, we pull more Pi bits (1024), but only 64-bits of the seed, to keep the API sane.
- The length of the input is not mixed into the AES block at the start to allow incremental construction, when the final length is not known in advance.
- The vector-loads are not interleaved, meaning that each byte of input has exactly the same weight in the hash. On the implementation side it requires some extra shuffling on older platforms, but on newer platforms it can be done with “masked” loads in AVX-512 and “predicated” instructions in SVE2.
Fun fact: AES instructions on x86 and Arm do slightly different things, but once you compensate for that, the performance is still great. Running StringWa.rs on an Intel Sapphire Rapids core, we get:
| std::hash | Rs | 0.43 GiB/s | 3.74 GiB/s |
| xxh3::xxh3_64 | C, C++, Rs, Py… | 1.08 GiB/s | 9.48 GiB/s |
| aHash::hash_one | Rs | 1.23 GiB/s | 8.61 GiB/s |
| gxhash::gxhash64 | Rs | 2.68 GiB/s | 10.81 GiB/s |
| stringzilla::hash | C, C++, Rs, Py… | 1.84 GiB/s | 11.23 GiB/s |
Generating Random Strings
The same hashing AES primitives can easily be used for high-throughput generation of random strings at speeds far exceeding what std::random_device and std::mt19937 can do. That’s typically achieved in one of 3 different modes:
- CTR (Counter Mode)
- OFB (Output Feedback Mode)
- CFB (Cipher Feedback Mode)
The first is easily parallelizable, which can be handy if we want to scramble all RAM on our machine, so the choice was clear. But it’s still tricky if every nano-second counts.
The reason why some of the StringZilla logic exceeds memcpy speeds, is the use of non-temporal stores, which bypass the CPU caches and write data directly to RAM. This reduces cache pollution and improves energy-efficiency on IO-heavy workloads. So for PRNGs it makes a lot of sense, but the extra complexity of handling misaligned writes wasn’t worth it, if you want to keep the PRNG seed-able and reproducible! Even without those tricks, the results are impressive, compared to other Rusty options:
| getrandom::fill | 0.18 GiB/s | 0.40 GiB/s |
| rand_chacha::ChaCha20Rng | 0.62 GiB/s | 1.72 GiB/s |
| rand_xoshiro::Xoshiro128Plus | 2.66 GiB/s | 3.72 GiB/s |
| zeroize::zeroize | 4.62 GiB/s | 4.35 GiB/s |
| sz::fill_random | 17.30 GiB/s | 10.57 GiB/s |
52-bit Math for Bioinformatics?!
There is, however, a kind of hashing where I still avoid AES primitives in favor of modulo integer arithmetic, but with a twist!
USearch, my search engine, uses 40-bit integers to identify entries - a non-standard size chosen to balance memory efficiency with a large enough address space to fit 1 trillion of vectors on 1 machine. Continuing the trend of obscure integer sizes, StringZilla v4 uses 52-bit integers to compute MinHashes, or “Min-wise independent permutations Locality Sensitive Hashing”.
First of all, I wasn’t sure if MinHash is a big thing. I doubted it even more once I looked at available software. But the community pressure was too high, so I caved in.
Looking at a text document of length $T$ bytes, we can extract $T - N + 1$ N-grams of length $N$. For example, the string “hello” contains the following 3-grams: “hel”, “ell”, and “llo”. Then, MinHash defines $D$ different hash functions $h_1, h_2, …, h_D$. For each hash function $h_i$, we compute $T - N + 1$ hashes - one for each N-gram - and take the minimum value:
$$ \text{MinHash}(h_i) = \min_{j=1}^{T - N + 1} h_i(\text{N-gram}_j) $$
Those $D$ minimum hash values form the MinHash signature of the document, a $D$-dimensional vector. Each of those hashes becomes more computationally expensive as the $N$ increases. So the overall complexity of computing the MinHash signature is $O(D \cdot (T - N) \cdot N)$. That’s a lot!
To reduce the complexity to $O(D \cdot (T - N))$, one can switch to Rabin-style rolling hashes, which I was doing in StringZilla v3 as well, but it wasn’t enough! Computing high-quality signatures required good enough intermediate hashes, either:
- a simple mixing scheme with larger 64-bit representations of hasher states, or
- a more complex mixing scheme with smaller 32-bit representations of hasher states.
But the third option is always the best one, right? As many of us, including readers of this blog and Less Slow C++, using float-s to do the dirty integer work is a common trick for tiny integers. What everyone forgets is that double-s are also a thing, and they have 53 bits of precision (52 stored bits plus 1 implicit bit). So if we can fit our hasher state in 52 bits, we can do all the mixing and modulo arithmetic using double-s, and then store the final result as a 32-bit integer.
| 1 | uint32_t | uint32_t | ★☆☆ | ★★★ | ★★☆ |
| 2 | uint64_t | uint64_t | ★★★ | ★★☆ | ★☆☆ |
| 3 | double | uint32_t | ★★★ | ★★☆ | ★★☆ |
Modern vectorized CPUs with 512-bit wide FMA units are surprisingly good at this, but GPUs are even better! It took a while to get rid of all the edge cases and ensuring the CPU and GPU kernels report the same fingerprints. At the end of the day:
- single-threaded Rust code: 0.5 MiB/s.
- H100 CUDA code: 392.37 MiB/s.
In 2023, Nvidia also added MinHash to their nvtext:: package. It uses the MurmurHash3_32 algorithm, as a foundation, and is somewhat more similar in design to the Rust’s probabilistic_collections package. Comparing these solutions purely on throughput isn’t fair, as the quality of the produced signatures matters and differs a lot.
| Serial MinHash for <ByteGrams> | 0.44 MiB/s | 0.47 MiB/s |
| 92.81% collisions | 94.58% collisions | |
| 0.8528 entropy | 0.7979 entropy | |
| pc::MinHash<ByteGrams> | 2.41 MiB/s | 2.37 MiB/s |
| 91.80% collisions | 93.17% collisions | |
| 0.9343 entropy | 0.8779 entropy | |
| szs::Fingerprints on 1x CPU | 0.56 MiB/s | 0.51 MiB/s |
| szs::Fingerprints on 16x CPUs | 6.62 MiB/s | 8.03 MiB/s |
| szs::Fingerprints on 1x GPU | 102.07 MiB/s | 392.37 MiB/s |
| 86.80% collisions | 93.21% collisions | |
| 0.9992 entropy | 0.9967 entropy |
Within StringWa.rs, I only currently look at the collision rate of individual dimensions across the dataset and the entropy of the bit distribution within signatures. The entropy is somewhat absolute, but the collision rate can only be compared within the same dataset. The ultimate benchmark, is of course, the quality of the nearest-neighbor search results, but that’s a topic for another post and a longer list of Information Retrieval techniques.
Sorting & Batch Operations
Most sorting algorithms are comparison-based, meaning that they rely on a series of comparisons between elements to determine their order. Comparing two integers is a single CPU instruction, but comparing two strings is much more complex. It involves a loop over the characters of both strings, comparing them one by one until a difference is found or the end of one string is reached:
In older StringZilla versions I’ve shown that the trivial optimization of sorting integer-represented prefixes before the rest of the string can yield significant speedups. The implementation was, however, a proof of concept, but now it’s a lot more usable and yields even better results:
| std::sort_unstable_by_key | 54.35 M comparisons/s | 57.70 M comparisons/s |
| rayon::par_sort_unstable_by_key on 1x CPU | 47.08 M comparisons/s | 50.35 M comparisons/s |
| arrow::lexsort_to_indices | 122.20 M comparisons/s | 84.73 M comparisons/s |
| sz::argsort_permutation | 182.88 M comparisons/s | 74.64 M comparisons/s |
It definitely still has room for improvement, and will pave the way to a broader range of batch-processing operations. Now that I have a fast enough thread-pool implementation, scaling will be easier!
Try Yourself
Beyond the algorithmic innovations, the engineering challenge was shipping across platforms and languages:
- For Python bindings alone, I ship to PyPI more platform-specific wheels per release than NumPy…
- And unlike NumPy, my libraries ship their own SIMD and GPGPU kernels, that need to be re-compiled for each platform, instead of relying on C interfaces of existing BLAS libraries…
- And unlike NumPy, StringZilla also ships for C, C++, CUDA, Rust, JavaScript, Go, and Swift, which all have their own peculiarities…
… all from the same repository’s CI on a free-tier GitHub Actions plan. So I won’t be surprised if there is still some weird issue propagating build flags and inferring a weird SIMD capability support on some platforms. To check if it works as expected on your end, for Python:
To check for detected capabilities:
For other programming languages, refer to the README.md and if something doesn’t work - share your issues on this thread on GitHub. For Rust, however, stuff should work fine, as its been used extensively for the StringWa.rs that you can pull and reproduce on your own hardware:
This work moves on small pushes from the community: a minimal repro, a perf trace, or a note to keep going. My thanks to everyone who contributed!
Special thanks to Nebius, my most-used cloud, for dependable, long-haul GPU capacity powering both personal experiments like this and Unum’s open-source work, from pre-training new perception & generative architectures to this kind of performance engineering.
Spread the word if you’d like to see more such open-source work 🤗
https://t.co/KbIf9uMQLa on GPUs: Databases & Bioinformatics 🦠
Levenshtein & NW/SW scores at 500+ GigCUPS
MinHash sketching in 52-bit integers at 300+ MB/s
More CPU-port-level optimizations & SVE2
A massive post on my largest open-source release of '25https://t.co/ZOXqqzZint
.png)






